HiPiler
Description
HiPiler is an interactive visualization interface for the exploration and visualization of regions-of-interest such as loops in large genome interaction matrices. Genome interaction matrices approximate the physical distance of pairs of genomic regions to each other and can contain up to 3 million rows and columns with many sparse regions. Traditional matrix aggregation or pan-and-zoom interfaces largely fail in supporting search, inspection, and comparison of local regions-of-interest (ROIs). ROIs can be defined, e.g., by sets of adjacent rows and columns, or by specific visual patterns in the matrix. ROIs are first-class objects in HiPiler, which represents them as thumbnail-like “snippets”. Snippets can be laid out automatically based on their data and meta attributes. They are linked back to the matrix and can be explored interactively. The design of HiPiler is based on a series of semi-structured interviews with 10 domain experts involved in the analysis and interpretation of genome interaction matrices. We describe six exploration tasks that are crucial for analysis of interaction matrices and demonstrate how HiPiler supports these tasks. We report on a user study with a series of data exploration sessions with domain experts to assess the usability of HiPiler as well as to demonstrate respective findings in the data.
Gallery
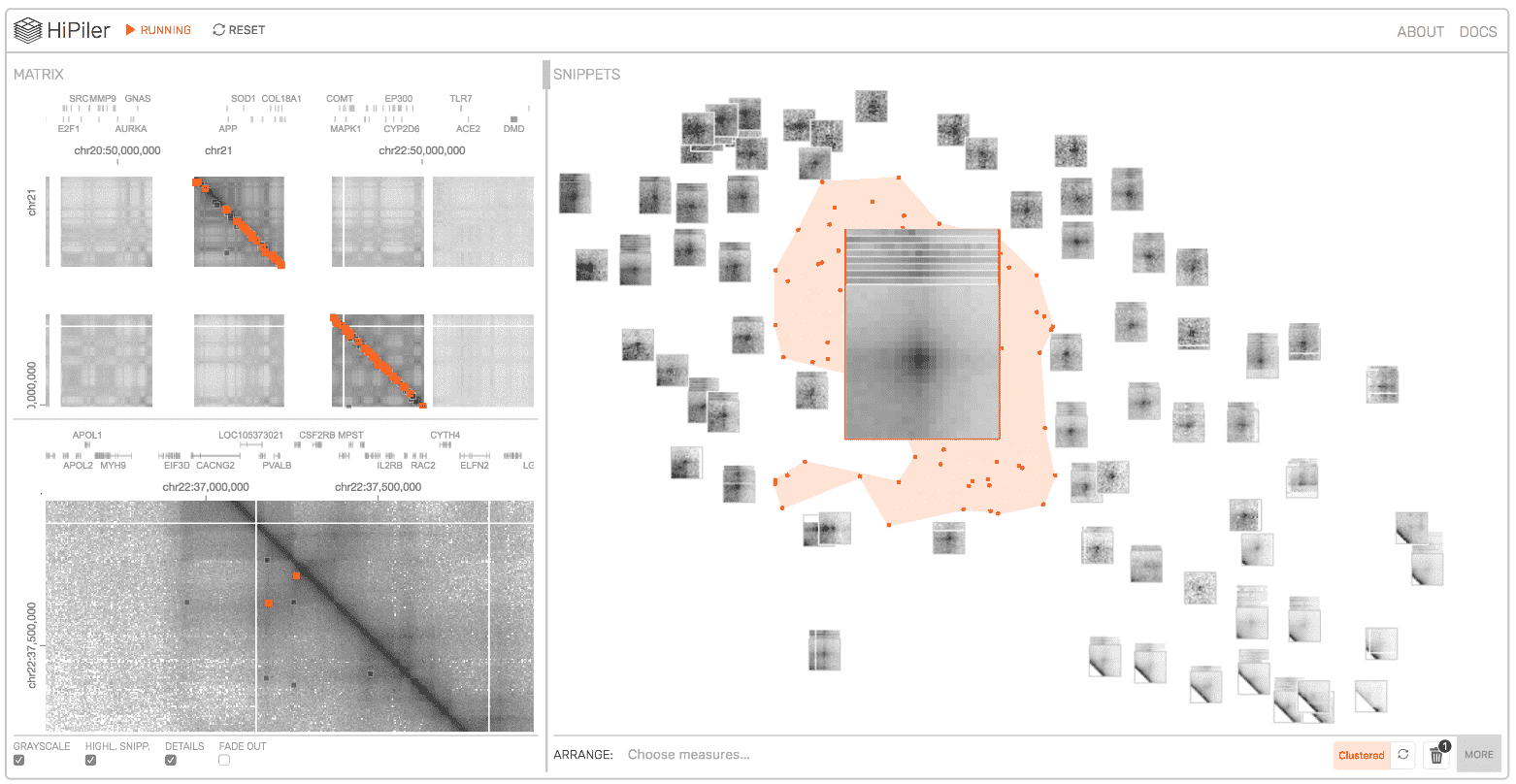 Screenshot
Screenshot

Publications
Websites
Media
- Nature News (2017) - TechBlog: HiPiler simplifies chromatin structure analysis
- "Using bioinformatic algorithms, researchers can query chromatin-contact datasets to find all such structures. But these algorithms may return thousands of hits. How is a researcher to study them? Enter HiPiler. Developed by Gehlenborg’s lab in collaboration with the lab of Hanspeter Pfister at the John A. Paulson School of Engineering and Applied Sciences at Harvard, HiPiler excises these features and presents them on a canvas as miniature snippets — basically tiny segments of the full-sized contact map, which users can then organize, sort, and cluster based on parameters such as noise or location."
Software
GitHub
- HiPiler Application
-
Clodius
A tool to tile 1D and 2D genomic data sets for use with HiGlass and HiPiler.
Funding
- 4D Nucleome Network Data Coordination and Integration Center (NIH U01CA200059)
National Institutes of Health - Common Fund - Visualization of (Epi)Genomic Data for Discovery of Disease-Associated Variants (NIH R00HG007583)
National Institutes of Health - NHGRI